| Version 9 (modified by lah, 10 years ago) (diff) |
|---|
Owl Processing Guide
The Owl thermal hyperspectral sensor is new to the arsf and so new problems may occur during processing. Please add any new issues to the bottom of this wiki. Details on the Owl sensor are available here and information on Owl testing is available here.
File Naming
Our scripts require that data be named according to the standard convention e.g. OWL219b-14-1, where 219 is the julian day, b the sortie, 14 the year and 1 the flightline. The files should have been renamed during unpacking, but if this is not the case, run proj_tidy.sh -e -p <project directory> to generate the move commands. Check that every move command is present before executing, as partial renaming cannot be completed in a second run. Taking a backup of the data files is recommended before renaming.
Dark Frames
The radiometric calibration will fail if the dark frames are saved in a separate file to the data (Ops sometimes do this). Check the capture directory for any extra files labelled like they might be dark frames and append them to the end of the data file. Remember to add the autodarkstartline key to the header file. Alternatively use the script stitchOwl.py.
Radiometric Calibration
Specim have provided a tool to process the Owl raw data into calibrated data. Details on using this tool are located here if required.
To batch process the data use: processOWL.py -p <project directory>
To process individual lines use: process_OWL_line.py -p <project directory> -f <flightline>
Each flightline should have it's own calibration data in the form of additional files prefixed T1 and T2 in the capture directory. If these are not present the script will produce a warning. Run the script again with the option -c auto once the submitted flightlines have produced the required calibration files (Check the log has passed the "Writing calibration file..." step), or manually specify a calibration file with -c <flightline with calibration data_calibration.rad>.
All output files are written to the flight line subdirectory of /processing/owl/flightlines/level1b and logs written to /processing/owl/logfiles. The output files consist of the data (*_proc.bil) and if not specified as inputs, the calibration (*_calibration.rad) and blinker files (*blinkers.dat), each with their own header file. During processing a symlink appears in the output flight line folder to prevent simultaneous processing over the same line. You may need to remove this if processing is aborted before it is automatically removed.
Files named as tp3f35cf3d_7182_4f17_8294_425fa4dc2953.raw are temporary files and will be removed on completion. If they remain check the logs for errors. No autodark found means the files need to be appended with dark frames.
Blinking Pixels/Vertical Stripes
During the radiometric calibration blinking pixels are tested for and removed from the data. It is likely that the default settings will not remove all the blinking pixels and the data will have vertical stripes. To remove these reprocess with a lower value of pixstabi e.g.
process_OWL_line.py -p <project directory> -f <flightline> -m pixstabi=0.005
The pixstabi modifier defines the standard deviation of the time series allowed for a normal pixel, so reducing this removes more pixels above this range. It is not expected that all blinking pixels can be removed as the blinking pixel routine does not yet appear to be optimal. Do not make the value too low as this will remove too many pixels and result in thick vertical stripes where the data has been replaced. It will also remove a considerable amount of spectral information, so check that the spectra still look reasonable. Somewhere between the first 2 images below is desirable.
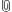
|
| Level 1b Owl data (2014 238) calibrated with different values of pixstabi (default 0.011, 0.005, 0.003, 0.0025, 0.002) |
Dropped Frames
If there are dropped frames they should be recorded in a .log file in the capture directory. These dropped frames need to be inserted into the level 1b data file as empty lines before geocorrection to map the flight line correctly.
Geocorrection
Create a dem in exactly the same way as for fenix. This should be saved in the processing/hyperspectral/dem directory, not the owl directories, so only 1 copy is required. e.g. from the project directory type create_apl_dem.py --aster.
Geocorrection is conducted in the same way as for fenix data, using APL. Generate a config file using generate_apl_config.py -o processing/owl/o2014219b.cfg (make sure it begins with an o) to keep processing separate from fenix. Currently you need to add the boresight values to the config file ( e.g. owl_boresight = 0.12 -0.32 0.32). You may want to delete or set to false the fenix lines if only processing owl data. Make sure the project directory structure is complete (same as fenix). Submit to the grid using specim_qsub.py <config_file> and calculate the SCTs. Once you have done this you can generate the mapped data (102 bands) by setting owl_bandlist = ALL and setting the SCT value for each line.
Early owl files have corrupt nav files, so if you get this message in the log or 'Requested sync time index is out of bounds in GetSyncDelay()' set the config file to use_nav = false.
If there appears to be stripping across the mapped files, bright and dark lines, or sections of a flight line don't match up the sensor has probably dropped frames. These can be checked in the .log file in the capture directory. These empty lines need to be inserted into the level 1b data file before geocorrection to map the flight line correctly.
Making a delivery
This currently needs to be done manually and there are no data quality reports to add yet.
Problems
There are likely to be problems with this newly developed processing chain. Please add problems (and solutions!) below and they will be fixed in due course.
- sctstart and sctend override owlsctstart and owlsctend in the config file.
- The boresight needs entering into the config file / needs adding to table.
- Bad pixels remain - will eventually look into a masking routine &/ get a better blinker detection routine from Specim